Introducing the SCIEX Citrine® MS/MS Medical Diagnostic (MD) mass spectrometer
In the modern diagnostic lab, analytical challenges demand increased sensitivity, speed, robustness and reliability of any diagnostic system, and mass spectrometry is no different. Designed and manufactured with industry-leading technologies, Citrine meets these challenges head on, giving confidence in results and the best possible service to patients. Citrine® MS/MS from SCIEX provides the ultimate performance and reliability to tackle today’s difficult assays, and the versatility to address tomorrow’s challenges. Delivering the legendary robustness and reliability of a SCIEX mass spectrometry solution, the Citrine® MS/MS system is specifically designed to meet the demands of clinical labs that require maximum sensitivity, high throughput, a wide dynamic range, and simplified sample preparation.
Sensitivity
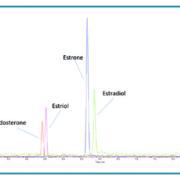
The Citrine® MS/MS – our most sensitive tandem mass spectrometer ever – provides the lowest possible limits of quantification, enabling the measurement of trace levels of biomarkers and metabolites at single-unit pmol/L concentrations. While sensitivity is key for accurate quantification, the enhanced sensitivity of Citrine can also allow streamlining of sample preparation, reducing consumables and reagent costs. (Figure 1)
Flexibility
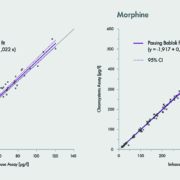
Getting the most from a single extraction and injection is clearly paramount to achieving increased efficiancies with any mass spectrometry analysis, and diagnostics is no different. With its fast MRM scanning (1 msec dwell times) and fast polarity switching (5 msec) between positive and negative ionization modes, the ability to analyse very large panels of compounds, across multiple compound classes is provided. (Figure 2)
A quantitative instrument… but so much more
The versatile Citrine® MS/MS system offers ESI and APCI ionization options, an extended mass range up to m/z 2000, and a large linear dynamic range, making this the perfect tool for the measurement of a large variety of polar and non-polar biomarkers and metabolites in biological fluids, over a large range of concentrations. Also available with SCIEX’s Triple Quadrupole Linear Ion Traps (QTRAP) technology, Citrine® becomes a hybrid triple quadrupole/linear ion trap mass spectrometer – a unique, flexible MS/MS system that can accommodate a wide variety of both quantitative and qualitative LC-MS/MS workflows. It is the ability to use both triple quadrupole and linear ion trap scan functions on a single platform – and even within a single LC-MS/MS run – that makes the QTRAP system adaptable to a wide variety of both screening and quantitative tests. On the quantitation side, in some cases isobaric interferences cannot be differentiated by MRM alone, since the interferences may have the same exact mass as the target compound. In these cases, the ability to use second-order fragmentation (MS/MS/MS, or MRM3) provides highly specific measurements and can remove chromatographic interferences caused by isomers and background ions, without the need for extended chromatography and reduced throughput. (Figure 3)
Legendary robustness and reliability
In the busy diagnostic lab, samples come in all flavours! Whatever matrix, whatever extraction – Citrine® delivers accurate and reliable results, day after day, time after time.
Citrine® MS/MS – truly the one solution for every challenge
The technologies within Citrine® provides cinical labs with a powerful diagnostic mass spectrometer that enables them to:
• Leverage the ultimate sensitivity of the Citrine® MS/MS system to reliably measure at picomole levels for clinically relevant biomarkers and metabolites
• Monitor 100’s of MRM transitions per analysis with uncompromised accuracy, precision and sensitivity
• Experience faster than ever data acquisition with 5 msec polarity switching
• Perform qualitative and quantitative analysis in a single injection with QTRAP® technology
• Enjoy the confidence provided by a medical device that meets the high quality and safety standards required by FDA regulations.
AB Sciex is doing business as SCIEX.
For in vitro diagnostic use.
Not available in all countries.
For more information: www.sciex.comclinical@sciex.com