Recent advances and perspectives in the molecular diagnosis of pneumonia
Despite some limitations, current molecular diagnostic methods have a great potential to include targets useful in the rapid identification of microorganisms and antimicrobial resistance, to analyse directly unprocessed samples and to obtain quantitative results in pneumonia, an entity of complex microbiological diagnosis due to the features of the pathogens commonly implicated.
by Dr A. Camporese
A change in culture without culture?
Developing accurate methods for diagnosing respiratory tract infections has long been a challenge for the clinical microbiology laboratory [1].
The current semi-quantitative agar-plate based culture method used in most clinical microbiology laboratories for analysing specimens from patients with suspected community-acquired pneumonia (CAP), hospital-acquired pneumonia (HAP), or ventilator-associated pneumonia (VAP), although adequate for recovering and identifying a wide variety of bacterial species from respiratory specimens, is slow, and cannot differentiate between colonization and infection. Moreover, results may be misleading, particularly if a Gram stain is not performed in parallel to ascertain the adequacy of expectorated sputum samples or endotracheal aspirates [2].
As the Infectious Diseases Society of America (IDSA) and American Thoracic Society (ATS) CAP guideline notes, one of the problems with diagnostic tests for respiratory tract infections “is driven by the poor quality of most sputum microbiological samples and the low yield of positive culture results” [3]. Moreover, the highest predictive value of a culture occurs only when Gram stain shows a predominant morphotype, and the culture yields predominant growth of a single recognized respiratory pathogen of that morphotype (e.g., Streptococcus pneumoniae) [2].
Unfortunately, such concordance decreases rapidly when specimens are collected after the initiation of antimicrobial therapy or when their arrival at the microbiology laboratory is significantly delayed.
One approach that may improve the diagnosis of respiratory tract infections and shorten the time necessary to place patients on appropriate therapy is the use of nucleic acid amplification methods.
Straight ahead toward molecular assays
Today clinical microbiologists appear to be on the threshold of a potentially important transition, with a substantial increase in the use of molecular diagnostic tests to replace or augment the century-old methods of culture, as many experts now view traditional microbiology as slow and antiquated, especially when compared with newer technologies used in other areas of laboratory medicine [4].
Traditional methods demonstrated poor sensitivity and specificity for detecting specific pathogens, particularly when the specimen being cultured is from a non-sterile anatomical compartment, such as the respiratory tract.
For this reason, molecular methods are becoming more widely used also for the detection of respiratory pathogens, in part because of their superior sensitivity, relatively rapid turnaround time, and ability to identify pathogens that are slow growing or difficult to culture.
However, to have a positive impact on patient management, molecular tests will need to be easy to use, and provide clear, definitive results that will give physicians the data necessary to start, or in some cases withhold, antimicrobial agents [5].
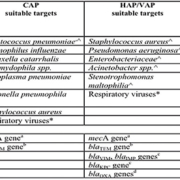
Further, to be really successful, industry must determine which combination of molecular targets [Table 1] and clinical specimens will produce results that will effectively guide anti-infective therapy regimens for patients with pneumonia or other respiratory tract diseases.
Another key challenge for industry will be to develop assays that are not only rapid, but also readily accessible, because development of an assay that is rapid, but unavailable on evening or night shifts, or at weekends, because of its technical complexity, limits the clinical value of the test.
Moreover, to be successful, molecular assays will need to be perceived by health care systems as cost-effective, but cost-effectiveness should be determined not only by comparison to the costs of performing slower, conventional methods in the laboratory, but also by consideration of the cost savings achieved from optimized antimicrobial therapy, decreased use of additional diagnostic tests, and shorter hospital stays [2].
To address issues on these topics, the IDSA and the Food and Drug Administration (FDA) co-sponsored a workshop on molecular diagnostic testing for respiratory tract infections in November 2009, with the participation of the FDA, industry, authorities in microbiology, statisticians and others. Respiratory tract infections were selected because this is the site of most infections treated with antibiotics in paediatric and adult practice, and they also represent a group of infections in which an etiologic agent is seldom identified in non-research settings [4].
The IDSA believes that patient care could be improved by accurate and rapid identification of pathogens, which would promote more judicious use of antibiotics, permit pathogen directed therapy, and provide potentially important
epidemiologic information.
Thus, the IDSA strongly desires development and implementation of molecular diagnostic tests that are easy, rapid, technically uncomplicated, applicable to specimens that are readily obtained, reasonably priced, sensitive and specific, because such tests will greatly improve antimicrobial stewardship, thereby helping to reduce the spread and impact of antibiotic resistance. Such tests will also facilitate conduct of clinical trials supporting the approval of new antibacterial agents [4].
Respiratory samples suitable for molecular assays
A variety of respiratory samples are amenable to molecular testing, including expectorated sputum, bronchoalveolar lavages (BALs), protected bronchial brushes, and endotracheal aspirates [2, 5, 6]. Of these, expectorated sputum samples are by far the most common respiratory samples submitted to the clinical microbiology laboratory, but are also the poorest in overall quality.
Endotracheal aspirates from ventilated patients are often of better quality than that of expectorated sputum obtained from patients with CAP/HAP, but may still be contaminated with upper respiratory tract flora.
Therefore, obtaining specimens from the site of infection that are not contaminated with upper respiratory tract flora remains to date a real and constant problem. BALs and protected brush samples seem more likely to yield samples from the site of infection, but require significantly more effort to obtain, and thus offer a much smaller market for a new molecular test.
Moreover, there is a significant gap in our knowledge as to how well molecular tests for bacterial pathogens would perform on expectorated sputum samples, compared with performance on BALs or protected brush samples from the same patient collected within a similar period [2].
This knowledge gap is also a barrier to test development, because a molecular test that cannot be performed on expectorated sputum (given all the problems with specimen quality) may not have broad enough appeal among physicians to make it a financially viable product (from the industry perspective).
Technology perspectives
There are a wide array of emerging technologies for the detection and quantification of respiratory pathogens directly from clinical specimens. Some of these technologies, such as real-time PCR, have potential for high-throughput testing, and others will allow rapid near patient testing, but more studies are needed to fully determine their performance characteristics and determine their ideal clinical application [6,7].
Molecular assays may target either a single pathogen or multiple respiratory pathogens in a single assay. There are merits to both single-pathogen and multiplex approaches. Certain bacterial respiratory pathogens cause such distinct clinical syndromes that assays that target them individually still have clinical utility. These include already many organisms, such as Chlamydophila pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, or Bordetella pertussis [Table 1].
Moreover, some multiplex assays for respiratory tract disease already include many targets for a rapid diagnosis of CAP, HAP, and VAP, but, in designing new assays, it will be critical to understand whether an assay for a determined number of bacterial pathogens will meet physicians’ needs and provide adequate data for initiating or altering anti-infective therapy [7, 8].
Potential and currently available targets for multiplex or individual molecular assays for respiratory tract samples in immunocompetent and/or immunocompromised patients with CAP, HAP, or VAP are presented in Table 1 [7].
Further, in this age of multidrug resistance, expanding the target selection to include key antimicrobial resistance genes that would alter existing therapy or guide empirical therapy, should also be considered [Table 1].
Lastly, if molecular-based diagnostic methods currently available are helpful in detecting single and multiple bacterial pathogens simultaneously, including the most frequent cause of CAP/HAP/VAP, the real-time PCR is also well known for its ability to quantify targets.
Where available, the application of quantitative molecular tests for the detection of key pathogens, such as S. pneumoniae, both in sputum and in blood, defining a threshold for classification, such as a colonizer or as an invasive pathogen, might be relevant in CAP patients, mainly in whom antibiotic therapy has been initiated, and might be a useful tool for severity assessment [9, 10].
Conclusion
Significant progress exists on the development and improvement of molecular-based methods feasible to be applied to the diagnosis of lower respiratory tract infection.
Multiplex assays, user-friendly formats, results in a few hours, high sensitivity and specificity in pathogen identification, detection of antibiotic resistance genes and target quantification, among others, are some of the contributions of novel molecular-based diagnosis approaches.
Developing new molecular tests for other bacterial respiratory pathogens, particularly microorganisms that can be both asymptomatic colonizers and overt pathogens of the respiratory tract, detection of pathogens and new key antimicrobial resistance genes in unprocessed samples, and determination of the microbial load by quantitative multi-pathogen tests will be some of the future challenges of molecular diagnosis in CAP/HAP/VAP.
References
1. Bartlett JG. Decline in microbial studies for patients with pulmonary infections. Clin Infect Dis 2004; 39: 170–172.
2. Tenover FC. Developing molecular amplification methods for rapid diagnosis of respiratory tract infections caused by bacterial pathogens. Clin Infect Dis 2011; 52(S4): S338–S345.
3. Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007; 44(S2): S27–S72.
4. Infectious Disease Society of America (IDSA). An unmet medical need: rapid molecular diagnostics tests for respiratory tract infections. Clin Infect Dis 2011; 52(S4): S384–S395.
5. Caliendo AM. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis 2011; 52(S4): S326–S330.
6. Lung M and Codina G. Molecular diagnosis in HAP/VAP. Curr Opin Crit Care 2012; 18: 487–494.
7. Camporese A. Impact of recent advances in molecular techniques on diagnosing lower respiratory tract infections (LRTIs). Infez Med 2012; 4: 237–244.
8. Johansson N, Kalin M, Tiveljung-Lindell A, et al. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis 2010; 50: 202–209.
9. Werno AM, Anderson TP, Murdoch DR. Association between pneumococcal load and disease severity in adults with pneumonia. J Med Microbiol 2012; 61: 1129–1135.
10. Woodhead M, Blasi F, Ewig S, et al. Guidelines for the management of adult lower respiratory tract infections-Full version. Clin Microbiol Infect 2011; 17(S6): E1–E59.
The author
Alessandro Camporese MD
Clinical Microbiology and Virology Department
S. Maria degli Angeli Regional Hospital, Pordenone, Italy
E-mail: alessandro.camporese@aopn.fvg.it