Lipidomic profiling using liquid chromatography mass spectrometry
The past 15 years have shown a slow but steady expansion of the field of lipidomics, which pushes analysis beyond understanding of traditional lipids (HDL-C, LDL-C and triglycerides). This article aims to introduce readers to the world of lipidomics and the remaining hurdles involved in this exciting field.
By Matthew W..K. Wong and Dr Nady Braidy
Introduction: what is lipidomics and why is it useful?
Lipidomics is a relatively novel subfield of ‘omics’ designed to identify and quantify hundreds to thousands of individual lipids in a given biological sample. In fact, the term ‘lipidomics’ did not exist in literature data-bases before 2004 and in 2016 lipidomics still formed under 1.% of ‘omics’ publications [1]. Despite the more established omics fields of genomics and proteomics having a clean head start on lipidomics in development and use, there has nevertheless been a huge explosion of interest in lipidomics in recent times, which can provide alternative angles of attack to answer questions relating to the biochemical basis of health and disease.
Although traditional lipids such as low-density lipoprotein cholesterol (LDL-C), high density lipoprotein (HDL-C) and total triglycerides are routinely analysed from blood and their concentrations applied to inform patients of clinical outcomes, these analyses do not capture the true complexity of lipids. In 2005, LIPID Metabolites and Pathways Strategy (LIPID MAPS) established a classification system of lipids which divided them into eight major classes: fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, saccharolipids, polyketides, isoprenols, and sterols [2]. Within each class, lipid species vary considerably in their degree of saturation (number of double bonds), their fatty acyl chain lengths determined by number of carbons, and the polarity of the head group with respect to the hydrophobic tails. This enables complex permutations of lipids to exist, and it has been estimated that there are over 100.000 naturally occurring lipids, though only 40.000 have been formally identified to date.
The sheer complexity of the lipidome at the molecular level appears to suggest that there are unique and specific physiological roles for these molecules. Apart from energy storage and membrane structure, lipids also participate in cellular transport, interact with ion channels and can function as signalling molecules, especially in detergent-resistant regions known as lipid rafts. These lipid raft regions are known to anchor many transmembrane proteins and may therefore be important signalling hotspots [3]. Given the relevance of lipids to many physiological processes, not surprisingly, lipidomics has been applied to identify potential biomarkers relating to health and disease including metabolic syndrome [4], cancer, Alzheimer’s disease [1] and other neurodegenerative diseases [5]. For example, altered phospholipids and sphingolipids have frequently been implicated in Alzheimer’s disease, whereas triglyceride levels have been shown to be dysregulated in behavioural variant frontotemporal dementia [5].
Mining the complexity of analytical techniques
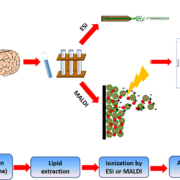
The main analytical tool of choice when it comes to lipid analysis is mass spectrometry (MS), where lipids are rapidly heated into the gas phase into charged ions, usually through soft ionization techniques such as electrospray ionization (ESI) [6]. Matrix assisted laser desorption/ionization (MALDI) can also be used on tissues, coupled with imaging MS to give an image of distribution intensity of various ions (Fig. 1). In ESI, the charged particles are injected into the mass spectrometer and suspended in helical motion about an orbitrap where the mass to charge ratio (m/z) of the analyte can be determined by assessing the angular momentum of the particle. Further information about the lipids of interest can be determined through experiments such as collision-induced dissociation and fragmentation of ions. Gas chromatography MS (GC-MS) is applied for fatty acid profiling, whereas liquid chromatography coupled MS (LC-MS) is particularly effective at detection of moderately polar lipids, such as glycerophospholipids. Lipidomic analysis can also be performed without chromatographic separation, in a technique known as direct infusion (or ‘shotgun lipidomics’) MS which takes into account differences in intrinsic ionization efficiency of lipid species according to their class, giving a broad fingerprint of the lipidome in a short amount of time [7]. This is a powerful and accurate technique as all lipids are analysed under the same conditions at the same time. LC-MS, however, first separates the lipids according to retention time. This way, major lipid classes are segregated on a time scale and lipids with similar m/z can be differentiated, enabling resolution of isobaric (lipids of different chemical composition but same mass) and isomeric (lipids with same chemical composition, but a different structure) lipid species which may be more difficult to resolve using direct infusion MS.
Quantitation
Quantitation of lipids can be achieved by comparing peak areas of lipids against internal standards (ISTDs). This can be either relative or absolute. ISTDs are routinely applied to correct for differences that occur owing to variation in experimental conditions, extraction efficiency, matrix effects and instrument performance, enabling the results of different LC-MS injection runs to be comparable [8].
All samples receive the same concentration of ISTDs applied consistently in the same batch, and the ISTDs are analysed simultaneously with the analytes of interest under the same experimental conditions.
For quantitation, ideally, each lipid species should have its own ISTD, but it is an expensive and time-consuming endeavour to account for tens to hundreds of lipids within the same class. Most commonly, only one or two ISTDs are applied for each lipid class. This approach requires ISTDs to have similar physio-chemical properties (ionization efficiency) to other lipids of the same class. It has been experimentally deduced that the number of carbons (chain length) and degree of saturation affect ionization efficiency to a smaller degree relative to the head group [8]. Most species of a lipid class sharing the same head group are expected to ionize similarly. In semi-quantitative analysis, where relative fold changes of lipids between groups are reported, a single ISTD per class is usually sufficient for normalization. The normalization is a simple process of determining the ratio of the analyte peak area to the corresponding internal standard peak area (Fig. 2a).
However, where more accurate and targeted quantitation is sought, multiple internal standards and calibration curves (external standards) are required (Fig. 2b). Choice of ISTD will vary depending on the analytes of interest. ISTDs should not be present endogenously in the sample and should have similar physicochemical properties. Low-physiologically occurring structural analogues, including odd chained lipids, or stable isotope-labelled standards are commonly used. ISTDs are synthesized and available for commercial use through manufacturers such as Avanti Polar Lipids, which now manufactures a cocktail of lipid standards to mimic concentrations found in plasma, called Avanti SPLASH Lipidomix.
Methodological concerns
This article focuses largely on application of lipidomics for plasma profiling, with venipuncture being relatively non-invasive and having the capacity for repeat collection. Lipidomics has been applied to the analysis of hundreds to thousands of individual samples [9] and through these experiments, researchers have identified important variables to take into consideration to maximize the available lipids for analysis. These include pre-analytical variables inherent in blood collection and storage. At the very least, lipids should be stored below −20.°C (even better at −80.°C) within 2.hours of collection, and freeze-thaw cycles should be kept at a minimum to prevent degradation of lipids. More detailed guidelines for blood collection, storage and attempts towards standardization of laboratory protocols have been reviewed [1, 10].
Further, the method of extraction will also determine the amount and type of lipids that can be analysed. For blood lipid extractions, the Folch and Matyash methods [11, 12] are considered gold standards and involve a biphasic extraction where lipids are suspended in the non-polar organic phase. More recently, a single-phase extraction method was introduced which bypasses the need to extract from the organic phase, with the entire set of lipids suspended in a single-phase supernatant [13]. Our laboratory has validated this method and confirmed that it clearly extracts lipids with as good, if not better efficiency compared to the Folch and Matyash methods [14]. Polar lipids are particularly well extracted with the single-phase method. Furthermore, the method demonstrated strong consistency, with median intra-assay and inter-assay coefficient of variation of 14.1.% and 14.4.%, respectively. Thus, repeated measurements within a batch and across batches separated over time yield consistent results and represent a strong alternative to the gold standard methods mentioned above.
Assessing the natural variation in lipids
Perhaps the greatest hurdle facing lipidomics research today is the lack of standard measurements. Although laboratory blood analysis routinely tests for concentrations of classical lipids such as LDL-C, HDL-C and TG, and standardized concentration ranges exist for diagnostic reference, no equivalent standardized tests exist for the rest of the lipidome. Without characterization of the range of baseline plasma concentrations of lipids within and between subjects it is more difficult to compare with disease states, where confounding variables can interfere with interpretation of results.
The Wenk group in the National University of Singapore has prioritized research on identi-fying intra-individual and inter-individual differences in the healthy human plasma lipidome. Their work has shown many lipids are regulated by circadian rhythms within individuals [15]; in addition, ethnicity may be another source of variation, where Chinese, Malay and Indian subjects had differences in their lipidomes [16]. Our laboratory has also shown age, sex, use of lipid lowering medications (such as statins) to be important determinants of lipid variation, in line with some previous studies [17]. Pre-analytical differences inherent in study design and sample characteristics must be considered well before lipidomic findings can be applied to the clinic. Further, NIST Standard Reference Material plasma has been used to estimate concen-tration ranges of various lipid classes [10]. The results suggest lipids in each plasma sample can vary by several orders of magnitude, and this means that no one analysis is able to capture all the lipids of interest. It is not unusual for multiple lipidomics platforms and extraction methods to be applied to overcome this setback – if time and resources permit.
Conclusion
Despite these hurdles, lipidomics continues to grow, driven by improvements in MS enabling much higher resolution detection and identification of lipids. As a greater under-standing of how lipids contribute to health and disease and how they are regulated by genetic and environmental factors develops, it is anticipated that in the near future, lipidomics will become more routinely applied towards identifying biologically important biomarkers for diagnostic and prognostic purposes.
References
1. Wong MW, Braidy N, Poljak A, Pickford R, Thambisetty M, Sachdev PS. Dysregulation of lipids in Alzheimer’s disease and their role as potential biomarkers. Alzheimers Dement 2017; 13(7): 810-827.
2. Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, et al. A comprehensive classification system for lipids. J Lipid Res 2005; 46(5): 839-861.
3. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science 2010; 327(5961): 46-50.
4. Meikle PJ, Wong G, Barlow CK, Kingwell BA. Lipidomics: potential role in risk prediction and therapeutic monitoring for diabetes and cardiovascular disease. Pharmacol Ther 2014; 143(1): 12-23.
5. Kim WS, Jary E, Pickford R, He Y, Ahmed RM, Piguet O, Hodges JR, Halliday GM. Lipidomics analysis of behavioral variant frontotemporal dementia: a scope for biomarker development. Frontiers in neurology 2018; 9: 104.
6. Brugger B. Lipidomics: analysis of the lipid composition of cells and subcellular organelles by electrospray ionization mass spectrometry. Annu Rev Biochem 2014; 83: 79-98.
7. Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 2005; 24(3): 367-412.
8. Wang M, Wang C, Han X. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why? Mass Spectrom Rev 2017; 36(6): 693-714.
9. Weir JM, Wong G, Barlow CK, Greeve MA, Kowalczyk A, Almasy L, Comuzzie AG, Mahaney MC, Jowett JB, et al. Plasma lipid profiling in a large population-based cohort. J Lipid Res 2013; 54(10): 2898-2908.
10. Burla B, Arita M, Arita M, Bendt AK, Cazenave-Gassiot A, Dennis EA, Ekroos K, Han X, Ikeda K, et al. MS-based lipidomics of human blood plasma: a community-initiated position paper to develop accepted guidelines. J Lipid Res 2018; 59(10): 2001-2017.
11. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957; 226(1): 497-509.
12. Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 2008; 49(5): 1137-1146.
13. Alshehry ZH, Barlow CK, Weir JM, Zhou Y, McConville MJ, Meikle PJ. An efficient single phase method for the extraction of plasma lipids. Metabolites 2015; 5(2): 389-403.
14. Wong MWK, Braidy N, Pickford R, Sachdev PS, Poljak A. Comparison of single phase and biphasic extraction protocols for lipidomic studies using human plasma. Front Neurol 2019; 10(879).
15. Chua EC, Shui G, Lee IT, Lau P, Tan LC, Yeo SC, Lam BD, Bulchand S, Summers SA, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci USA 2013; 110(35): 14468–14473.
16. Saw WY, Tantoso E, Begum H, Zhou L, Zou R, He C, Chan SL, Tan LW, Wong LP, et al. Establishing multiple omics baselines for three Southeast Asian populations in the Singapore Integrative Omics Study. Nat Commun 2017; 8: 653.
17. Wong MWK, Braidy N, Pickford R, Vafaee F, Crawford J, Muenchhoff J, Schofield P, Attia J, Brodaty H, et al. Plasma lipidome variation during the second half of the human lifespan is associated with age and sex but minimally with BMI. PLoS One 2019; 14(3): e0214141.
The authors
Matthew WK Wong BMedSci; Nady Braidy* BMedSci (Hons I Phys/Pharm), MPharm, DipInnovMan, GradCertResMan, PhD
Centre for Healthy Brain Ageing, School of Psychiatry, Faculty of Medicine, University of New South Wales, Sydney, New South Wales, Australia 2052
*Corresponding author
E-mail: n.braidy@unsw.edu.au
Figure 2 (b). Quantitation by internal standards. Absolute quantification involves setting up a calibration curve where various known concentrations of internal standards (ISTDs) and their corresponding peak areas are plotted, and a linear response obtained. The unknown concentration of lipid is then determined by taking the peak area to the calibration line and interpolating to the concentration axis.