Recent progress in host transcriptome profiling paves the way for a future point-of-care test for pediatric TB
A diagnostic point-of care (POC) test would greatly improve case detection and management of pediatric tuberculosis (TB). Herein, we provide a brief overview of the challenges associated with diagnosing TB in children and recent evidence that points towards a future POC test based on transcriptomes in peripheral blood.
by Dr S. Jenum, Dr J.E. Gjøen, R. Bakken, Dr D. Sivakumaran and Prof. H.M.S. Grewal
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb), is a leading cause of childhood death and morbidity worldwide estimated to cause 1 million new cases in children <15 years of age in 2014 [1]. The gold standard for a diagnosis of pulmonary TB is growth of Mtb from respiratory specimens. However, culture-confirmed TB only occurs in about 30% of pediatric TB cases because of the paucibacillary nature of the disease and the difficulties in obtaining representative specimens [2]. For these reasons, children have been perceived less contagious and, therefore, not considered a priority for case finding within national TB programmes. This has resulted in detection rates being as low as 35% [3], with evident consequences for childhood death and morbidity. An accurate diagnostic test that provides a rapid, sensitive and specific result, enabling early initiation of treatment at the point-of-care (POC), ‘a diagnostic POC test’, could prevent this. The declaration by the World Health Organization (WHO) of pediatric TB as a neglected area of science and the stated urgent need for improved diagnostics [4], has spurred the search for TB diagnostic biomarkers in children. We provide a brief overview of the challenges associated with diagnosing TB in children and recent evidence that points towards a future POC test based on transcriptomes in peripheral blood.
Diagnosing TB in children
Diagnostic approaches and criteria for pediatric TB have recently been extensively reviewed [5]. In summary, sampling of respiratory specimens for confirmation of diagnosis by culture is strongly recommended, but requires repeated sampling of induced sputum and gastric aspirates, which are cost and labour intensive [5]. Notably, as many as 70% of children are culture negative even under optimized study conditions [2]. Therefore, in most cases, a diagnosis is based on a diagnostic algorithm including clinical signs and symptoms, radiological findings suggestive of TB and immunological evidence of previous Mtb sensitization [5]. Clinical diagnostic algorithms have not being adequately validated [6], and radiological findings are often unspecific [5]. The tuberculin skin test (TST) and the newer interferon-γ release assays used as a surrogate for Mtb infection cannot differentiate between infection and disease [7], and have reduced sensitivity in young and malnourished children [8, 9], but may be complementary by improving sensitivity in such clinical contexts [5].
Requirements for a POC test
Direct microscopy of sputum smears looking for acid-fast bacilli, is widely implemented at primary diagnostic centres for the diagnosis of pulmonary TB in adults, but the sensitivity is highly variable (20–80%) [10]. Since 2010, the use of the Xpert MTB/RIF assay on sputum samples has expanded in low- and middle-income countries [11], serving as a POC test in adults. The assay is based on direct identification of Mtb in specimens by a nucleic acid amplification test and detects in children three times the cases detected by direct microscopy and ~70% of the cases detected by liquid culture [12]. The WHO recommends that a POC triage test, meant to narrow down the population that needs further diagnostic work-up, should achieve a sensitivity of 90% and a specificity of 70%. For a diagnostic POC test, the sensitivity should be >95% and the specificity >98% [13]. A POC test based on Mtb detection will never meet these requirements in children because of the high proportion of culture-negative pulmonary TB [2], as well as the significant proportion of extrapulmonary TB [5]. Host biomarkers reflecting the ongoing pathological processes resulting from Mtb infection may hold greater promise for this purpose. Moreover, peripheral whole blood (WB) is more easily available for diagnosis than respiratory or tissue samples, particularly in children. Therefore, biomarkers based on transcriptomes in WB have gained increasing interest.
Genome-wide analysis of RNA expression
A landmark study by Berry et al., based on genome-wide analysis of RNA expression in unstimulated WB from adults, identified an 86-transcript signature able to discriminate active TB from other inflammatory and infectious diseases [14]. This transcriptomic signature consisted mainly of neutrophil-driven interferon (IFN)-inducible genes, which until then, had not been considered essential players in TB immunity. Further, a decreased abundance of B-cell transcripts in TB patients [14], challenged the paradigm of B-cells and humoral immunity being of little importance in protection against Mtb. Following this study, the upregulation of type I IFN-inducible genes and altered expression of B-cell related genes has been confirmed in adults [15–19] and adolescents [20].
To our knowledge, two previous studies in children have been published based on genome-wide transcriptional profiling of WB in analysis of RNA expression in Warao Amerindians [21], and in three African cohorts [22]. Verhagen et al. addressed the diagnostic gap in discriminating TB disease from latent TB, and identified a 5-transcript signature with a prediction error of 11% in discriminating active from latent TB. The signature classified correctly 78% of TB cases and 100% of latent TB cases in a validation cohort comprising children with non-TB pneumonia [21]. Interestingly, the 5-transcript signature, when applied on transcriptional data-sets generated by microarray analyses of WB from adults performed similarly, whereas the gene signatures identified in these adults were useless in the cohort of Warao Amerindian children [21]. This questions the validity in extrapolating results from transcriptional TB biomarker research in adults to children, highlighting the importance of studies in pediatric populations.
Applying a more real-life diagnostic setting with inclusion of children evaluated for suspected TB applying a graded diagnosis of TB in line with the consensus statement [6], Anderson et al. identified a 51-transcript signature that distinguished TB from other diseases in South African and Malawian children, subsequently validated in Kenyan children. A risk score based on the signature identified confirmed TB with a sensitivity of 82.9% and a specificity of 83.6%, and discriminated between other disease and culture-negative TB with a sensitivity ranging from 35–82% [22].
A more cost-effective method for assessing transcriptomes
Genome-wide analyses of transcriptomes are costly and resource-intensive but can be seen as a necessary first step in identifying markers with potential for subsequent refinement as POC tests. A test more suitable for hypothesis-testing or more high-throughput screening of transcriptional signatures with relevance in TB, has recently been developed [23]. The dual-colour Reverse Transcriptase-Multiplex Ligation-dependent Probe Amplification (dcRT-MLPA) can rapidly profile multiple host genes with a dynamic range and accuracy comparable to real-time qPCR and RNA sequencing at a cost of €5–7 per sample [24], most likely simplifying the translation of findings into diagnostic tools in resource-poor settings. The dcRT-MLPA method has been applied in different studies assessing TB biomarkers [19, 23–25]. Recently, incorporating the results from unbiased gene-expression profiling, the gene panels adapted for the dcRT-MLPA platform have been expanded to include type I IFN-inducible genes as well as genes covering adaptive immunity, including B-cells [25].
The dcRT-MLPA method in the context of pediatric TB
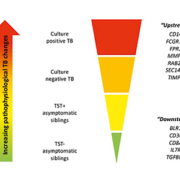
We have applied the dcRT-MLPA method to identify TB diagnostic transcriptome signatures in pediatric TB settings in India [26, 27]. We have also examined the immunological basis for discordant TST and Quantiferon® responses [28], as well as the potential interference of non-tuberculous mycobacteria on transcriptional biomarkers [29]. The younger the child, the more challenging the diagnosis of TB. We therefore explored transcriptional biomarkers in Indian children aged <3 years referred for a TB diagnostic work-up in a prospective cohort study of BCG-vaccinated neonates. We found that RAB33A alone discriminated between clinical TB and Mtb infection with an area under the curve (AUC) of 77.5%, whereas a 5-transcript signature effectively discriminated between clinical TB and controls (AUC 91.7%) [26]. Then, in a cohort of older Indian children (mean age ~9 years) with intrathoracic TB and their siblings, we assessed transcriptional biomarkers across the spectrum of TB disease. We identified 12 biomarkers consistently associated with clinical groups ‘upstream’ towards confirmed TB or ‘downstream’ towards a decreased likelihood of TB disease (Fig. 1), suggesting a correlation with Mtb-related pathology and high relevance to a future POC test. Furthermore, an 8-transcript signature separated children with TB from asymptomatic siblings (AUC 88%) [27]. Notably, these transcriptome signatures provided a superior sensitivity for confirmed TB compared with the Xpert MTB/RIF performed on gastric lavage [2]. Many of the biomarkers within these signatures have been confirmed in adults with Mtb infection or disease [19, 23, 24].
Future perspectives
We are currently exploring the performance of the extended dcRT-MLPA panels of genes which include type I IFN-signalling, myeloid cell activation, general inflammation and B-cell related genes in Indian children with intrathoracic TB. Transcriptome signatures identified in a discovery set consisting of randomly assigned TB cases and their healthy siblings will be validated in a diagnostic setting of symptomatic children aged <3 years.
We are also exploring the association between transcriptional biomarkers and the extent of TB disease and the outcome of TB treatment in children. The monitoring and evaluation of TB treatment is hampered by the limited tools available to guide decision-making during the long treatment period. Based on our unpublished results, as well as from findings in adults [16, 17, 23] we believe that treatment monitoring by means other than a negative culture result at 2 months post-treatment [30] will be possible.
Conclusions
Transcriptome-based tests meeting the requirements for both a screening and a diagnostic POC test would greatly improve case detection in children and subsequently reduce morbidity and mortality attributable to TB [13]. Further, a POC test capable of guiding treatment would prevent treatment failure and the subsequent emergence of resistant Mtb strains.
References
1. WHO. Global tuberculosis report 2015. ISBN 978-9241565059. http://apps.who.int/iris/bitstream/10665/191102/1/9789241565059_eng.pdf.
2. Dodd LE, Wilkinson RJ. Diagnosis of paediatric tuberculosis: the culture conundrum. Lancet Infect Dis. 2013; 13(1): 3–4.
3. Dodd PJ, Gardiner E, Coghlan R, Seddon JA. Burden of childhood tuberculosis in 22 high-burden countries: a mathematical modelling study. Lancet Glob Health 2014; 2(8): e453–459.
4. WHO. Roadmap for childhood tube culosis: towards zero deaths. WHO 2013;http://apps.who.int/iris bitst eam/10665/89506/1/9789241506137_eng pdf.
5. Perez-Velez CM, Marais BJ. Tuberculosis in children. N Engl J Med. 2012; 367(4): 348–361.
6. Graham SM, Ahmed T, Amanullah F, Browning R, Cardenas V, Casenghi M, Cuevas LE, Gale M, Gie RP, et al. Evaluation of tuberculosis diagnostics in children: 1. Proposed clinical case definitions for classification of intrathoracic tuberculosis disease. Consensus from an expert panel. J Infect Dis. 2012; 205 Suppl 2: S199–S208.
7. Pai M, Denkinger CM, Kik SV, Rangaka MX, Zwerling A, Oxlade O, Metcalfe JZ, Cattamanchi A, Dowdy DW, et al. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev. 2014; 27(1): 3–20.
8. Machingaidze S, Wiysonge CS, Gonzalez-Angulo Y, Hatherill M, Moyo S, Hanekom W, Mahomed H. The utility of an interferon gamma release assay for diagnosis of latent tuberculosis infection and disease in children: a systematic review and meta-analysis. Pediatr Infect Dis J. 2011; 30(8): 694–700.
9. Jenum S, Selvam S, Mahelai D, Jesuraj N, Cardenas V, Kenneth J, Hesseling AC, Doherty TM, Vaz M, Grewal HM. Influence of age and nutritional status on the performance of the tuberculin skin test and QuantiFERON-TB gold in-tube in young children evaluated for tuberculosis in Southern India. Pediatr Infect Dis J. 2014; 33(10): e260–269.
10. WHO. Report of the Tenth Meeting of the Strategic and Technical Advisory Group for Tuberculosis (STAG-TB) 2010. WHO 2010; http://www.who.int/tb/advisory_bodies/stag_tb_report_2010.pdf.
11. Qin ZZ, Pai M, Van GW, Sahu S, Ghiasi M, Creswell J. How is Xpert MTB/RIF being implemented in 22 high tuberculosis burden countries? Eur Respir J. 2015; 45(2): 549–554.
12. Nicol MP, Workman L, Isaacs W, Munro J, Black F, Eley B, Boehme CC, Zemanay W, Zar HJ. Accuracy of the Xpert MTB/RIF test for the diagnosis of pulmonary tuberculosis in children admitted to hospital in Cape Town, South Africa: a descriptive study. Lancet Infect Dis. 2011; 11(11): 819–824.
13. WHO. Global Tuberculosis Report. WHO 2014; http://apps.who.int/iris/bitst eam/10665/137094/1/9789241564809_eng.pdf?ua=1.
14. Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466(7309): 973–977.
15. Ottenhoff TH, Dass RH, Yang N, Zhang MM, Wong HE, Sahiratmadja E, Khor CC, Alisjahbana B, van Crevel R, et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One 2012; 7(9): e45839.
16. Bloom CI, Graham CM, Berry MP, Wilkinson KA, Oni T, Rozakeas F, Xu Z, Rossello-Urgell J, Chaussabel D, et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One 2012; 7(10): e46191.
17. Cliff JM, Lee JS, Constantinou N, Cho JE, Clark TG, Ronacher K, King EC, Lukey PT, Duncan K, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis. 2013; 207(1): 18–29.
18. Kaforou M, Wright VJ, Oni T, French N, Anderson ST, Bangani N, Banwell CM, Brent AJ, Crampin AC, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 2013; 10(10): e1001538.
19. Sutherland JS, Loxton AG, Haks MC, Kassa D, Ambrose L, Lee JS, Ran L, van Baarle D, Maertzdorf J, et al. Differential gene expression of activating Fcγ receptor classifies active tuberculosis regardless of human immunodeficiency virus status or ethnicity. Clin Microbiol Infect. 2014; 20(4): O230–238.
20. Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 2016; 387(10035): 2312–2322.
21. Verhagen LM, Zomer A, Maes M, Villalba JA, Del Nogal B, Eleveld M, van Hijum SA, de Waard JH, Hermans PW. A predictive signature gene set for discriminating active from latent tuberculosis in Warao Amerindian children. BMC Genomics 2013; 14: 74.
22. Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, Crampin AC, Dockrell HM, French N, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. New Eng J Med. 2014; 370(18): 1712–1723.
23. Joosten SA, Goeman JJ, Sutherland JS, Opmeer L, de Boer KG, Jacobsen M, Kaufmann SH, Finos L, Magis-Escurra C, et al. Identification of biomarkers for tuberculosis disease using a novel dual-color RT-MLPA assay. Genes Immun. 2012; 13(1): 71–82.
24. Haks MC, Goeman JJ, Magis-Escurra C, Ottenhoff TH. Focused human gene expression profiling using dual-color reverse transcriptase multiplex ligation-dependent probe amplification. Vaccine 2015; 33(40): 5282–5288.
25. Sloot R, Schim van der Loeff MF, van Zwet EW, Haks MC, Keizer ST, Scholing M, Ottenhoff TH, Borgdorff MW, Joosten SA. Biomarkers can identify pulmonary tuberculosis in HIV-infected drug users months prior to clinical diagnosis. EBioMedicine 2015; 2(2): 172–179.
26. Dhanasekaran S, Jenum S, Stavrum R, Ritz C, Faurholt-Jepsen D, Kenneth J, Vaz M, Grewal HM, Doherty TM; TB Trials Study Group. Identification of biomarkers for Mycobacterium tuberculosis infection and disease in BCG-vaccinated young children in Southern India. Genes Immun. 2013; 14(6): 356–364.
27. Jenum S, Dhanasekaran S, Lodha R, Mukherjee A, Kumar Saini D, Singh S, Singh V, Medigeshi G, Haks MC, et al. Approaching a diagnostic point-of-care test for pediatric tuberculosis through evaluation of immune biomarkers across the clinical disease spectrum. Sci Rep. 2016; 6: 18520.
28. Dhanasekaran S, Jenum S, Stavrum R, Ritz C, Kenneth J, Vaz M, Doherty TM, Grewal HM; TB Trials Study Group. Concordant or discordant results by the tuberculin skin test and the quantiFERON-TB test in children reflect immune biomarker profiles. Genes Immun. 2014; 15(5): 265–274.
29. Dhanasekaran S, Jenum S, Stavrum R, Wiker HG, Kenneth J, Vaz M, Doherty TM, Grewal HM; TB Trials Study Group. Effect of non-tuberculous Mycobacteria on host biomarkers potentially relevant for tuberculosis management. PLoS Negl Trop Dis. 2014; 8(10): e3243.
30. Wallis RS, Kim P, Cole S, Hanna D, Andrade BB, Maeurer M, et al. Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect Dis. 2013; 13(4): 362–372.
The authors
Synne Jenum*1 MD, PhD; John Espen Gjøen2 MD, Rasmus Bakken2; Dhanasekaran Sivakumaran2 PhD; Harleen M.S. Grewal2 MD, PhD, DTMH
1Department of Infectious Diseases, Oslo University Hospital, Pb 4950, N-0424 Oslo, Norway
2Department of Clinical Science, University of Bergen, Pb 7804, N-5021 Bergen, Norway
*Corresponding author
E-mail: synnejenum@gmail.com