Chest pain and elevated cardiac troponin: a typical case of myocardial infarction? Perhaps not…
Chest pain is a common presentation in Emergency Medicine and can cause a diagnostic dilemma. Does the patient need urgent admission for percutaneous coronary intervention following a myocardial infarction? Or will discharge with reassurance and a bottle of antacids sort out their indigestion? Measurement of cardiac troponin has become an essential part of emergency assessment of chest pain. Despite this, clinicians and laboratory professionals should be conscious that elevated cardiac troponin is not inevitably indicative of cardiac pathology – and in some cases, may be deceptive…
by Ceri Parfitt and Dr Christopher Duff
Troponin and assessment of acute chest pain
The Universal Definition of Myocardial Infarction and Injury has incorporated measurement of cardiac troponin (cTn) since 2000, with the most recent revision in 2012 [1]. In the UK, the National Institute of Health and Care Excellence (NICE) the ‘Chest pain of recent onset: assessment and diagnosis’ pathway [2] is well established in clinical practice, and Emergency Department clinicians are confident in the use of cTn assays to identify patients with acute coronary syndrome (ACS).
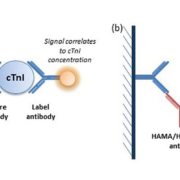
The troponin complex comprises three regulatory proteins – troponin C, troponin T and troponin I – essential for skeletal and cardiac muscle contraction, through regulation of actin/myosin filaments. Cardiac-specific isoforms of both troponin T (cTnT) and troponin I (cTnI) have been identified. cTn complexes can be detected in the blood within 2–3 hours of myocardial damage, peak within 24 hours and persist for 1–2 weeks [3, 4]. This characteristic pattern of release and specificity for cardiac injury has cemented cTn as the biomarker of choice for diagnosis of ACS, replacing creatinine kinase, aspartate transaminase and lactate dehydrogenase. Measurement of cTn is a standard assay available on an urgent basis in the majority of modern clinical laboratories. Both cTnT and cTnI can be measured by two-site ‘sandwich’ immunoassay, based on formation of complexes of cTn, a ‘capture’ anti-cTn antibody and a ‘label’ anti-cTn antibody (Fig. 1a). Owing to patent regulations, Roche Diagnostics is the sole distributor of cTnT immunoassays, whereas various companies offer cTnI immunoassays on several different platforms. Although results cannot be directly compared, the two assays largely provide the same diagnostic information.
A patient presents with chest pain and raised troponin
Despite the widespread use of cTn in Emergency Departments worldwide, clinicians may often be unaware of spurious causes of raised cTn. Application of the pathway without considering patient history and previous presentations can potentially lead to invasive, unnecessary (and expensive) investigations. We recently encountered an example of this in our local Emergency Department. A 34-year-old male with no significant cardiac risk factors repeatedly presented to the Emergency Department complaining of chest pain over a period of 3 years, resulting in more than 40 individual analyses of cTnI (using the Siemens TnI-Ultra assay on ADVIA Centaur XP). The results were variable, but always elevated (48–4030 ng/L, decision limit 40 ng/L). The patient underwent thorough investigation, but no cardiac pathology was ever identified. The Cardiology Department contacted the laboratory to discuss the apparent mismatch between patient presentation and biochemical findings.
Has there been a laboratory error?
Spurious results can arise for a number of reasons, including mislabelled samples, sample contamination, poor sample quality, analyser malfunction, calibration/quality control errors and transcription errors [5]. Where possible, these factors are minimized by stringent sampling techniques and robust laboratory procedures, or can be identified through the technical and clinical validation processes, and highlighted to the clinical team by laboratory staff. In many cases, a simple repeat sample is sufficient to confirm/rule out error. However, clinicians should always be vigilant for results that are not consistent with patient presentation. The persistent nature of the elevated cTnI in this case – over a period of 3 years – rules out sporadic errors, such as mislabelled samples or analyser malfunction. Furthermore, no other patients with unexpectedly high cTnI concentrations were discussed with the laboratory. These factors strongly suggested that the solution to the mystery lay within the patient, rather than in management of their samples.
Alternative causes of elevated troponin: cardiac and non-cardiac
Although elevated cTn is typically associated with myocardial infarction, other cardiac pathologies also cause cTn release into the circulation through direct or hypoxic myocyte damage. These include aortic dissection, pericarditis, myocarditis, acute heart failure and cardiac contusion following trauma [4, 6]. Unsurprisingly, cardiac interventions such as defibrillator shocks, coronary angioplasty, percutaneous coronary intervention and open heart surgery are also associated with variable increase in cTn [6]. Chemotherapy agents, including anthracyclines, alkylating agents, anti-metabolites and anti-microtubules are all associated with toxic damage to myocytes [4].
In addition to cardiac sources of cTn, non-cardiac sources must also be considered. A significant proportion (estimated at 36–85 %) of patients admitted to critical care units for sepsis/systemic inflammatory response syndromes (SIRS) have elevated cTn concentrations. In these patients, cTn release is thought to be a consequence of oxygen supply-demand mismatch in the myocardium: although oxygen demand is increased, due to fever and tachycardia, systemic hypoxemia reduces oxygen supply due to respiratory failure, circulatory dysfunction and hypotension [7, 8]. Patients with end-stage renal disease (ESRD) are often noted to have chronically elevated cTn. The underlying cause for this is unclear, but various hypotheses have been presented, including reduced renal clearance of cTn microfragments, sub-clinical cardiac disease, and metabolic abnormalities [9]. cTn concentrations are also found to be elevated in patients with acute pulmonary embolism. Again, the cause has not been conclusively established, but it is believed that abrupt increases in pulmonary artery pressure leads to dilatation of the right ventricle, with associated myocyte injury [4].
For most patients, any of these situations can be identified by examining the patient records, or through further cardiac investigations. In the case presented here, after thorough analysis of the patient’s history, and given the resolutely normal cardiac investigations, we concluded that it was unlikely that the elevation in cTn was due to pathological factors, whether cardiac or non-cardiac. This led us to examine the local cTnI assay – and the patient’s samples – in more detail, to ascertain the cause of the spurious results.
Could immunoassay interference be involved?
Non-pathological causes of false positives in cTn analysis include human anti-mouse antibodies (HAMA), rheumatoid factor (RF) and heterophilic antibodies [10–12]. HAMA are characterized by strong avidity to well-defined antigens. They are thought to develop following exposure to mouse immunoglobulins, which may occur through monoclonal antibody therapy, vaccination, infection, blood transfusion, or simply through animal handling [10]. In contrast, heterophilic antibodies are endogenous antibodies present in serum/plasma produced against poorly defined antigens. They often have weak affinity, but multiple specificities. Heterophilic antibodies may develop following infection and are thought to affect up to 30% of the population [10, 12, 13]. RFs are endogenous human antibodies with heterophilic activity, often arising following autoimmune disease [12].
Genuine assay interference is less commonly encountered these days, with the development of highly specialized commercial assays. Most modern immunoassays contain agents able to block low concentrations of interfering proteins. However, the fact that interference is less frequently observed may lead laboratory staff members to believe that interference is less prevalent in the general population. This is not the case. In fact, the prevalence of assay interference may be increasing, as immunotherapy and the use of radiolabelled antibodies are established in routine practice [13]. Although heterophilic antibodies usually have little clinical significance in vivo, their ability to interfere with two-site immunoassays can have major effects on patient management. The magnitude of interference varies between samples and may even vary within a patient over time [13]. The mechanism for antibody interference is complex and has not been fully elucidated, but is likely to involve ‘bridge’ formation between the capture antibody and label antibody (Fig. 1b) [13, 14]. Several standard techniques are available to investigate assay interference [12, 13]. In this case, we used dilution studies, polyethylene glycol precipitation, heterophilic antibody blocking tubes and an alternative assay method. In all cases, samples from the patient were compared with a control sample with a similar cTnI concentration, from a patient with confirmed ACS.
Investigations into immunoassay interference
Serial dilution of samples should produce a linear decrease in concentration if interfering species are absent. However, if interfering substances are present, then non-linear recovery following dilution is commonly observed, as the interferent is ‘diluted out’. Both the patient’s serum and control serum were diluted with saline, and cTnI was measured in each dilution. As expected, the cTnI concentration in the control sample decreased proportionally to the dilution factor. In contrast, any dilution of the patient sample resulted in undetectable cTnI, consistent with presence of an interfering species (Fig. 2a). Immunoglobulins can be removed by treatment with polyethylene glycol (PEG). PEG forms poorly soluble complexes with immunoglobulins, which can be precipitated then removed by centrifugation, before the sample is re-analysed. In the patient sample, PEG precipitation resulted in undetectable levels of cTnI. Levels were also reduced in a control sample (62% recovery), though it is not uncommon to see at least some reduction in recovery using this approach, regardless of whether interfering immunoglobulins are present or not (Fig. 2b). Heterophilic blocking reagents prevent non-analyte mediated bridging of antibodies by heterophilic interference. Incubation of a patient sample in a heterophilic blocking tube (Scantibodies Laboratory Inc.) before cTnI analysis resulted in 4% recovery of cTnI, suggesting analytical interference. In the control sample, 94% of cTnI was recovered (Fig. 2c). Finally, a patient sample was sent to a referral laboratory for analysis of cTnT, as opposed to cTnI. A cTnT concentration below the decision limit (99th percentile) was obtained (cTnI result: 398 ng/L; cTnT result: 6 ng/L) (Fig. 3).
As initial investigations strongly suggested the presence of an assay interferent, further work was conducted to identify the precise nature of the interfering species. Two patient samples with elevated cTnI were analysed for presence of HAMAs, which was negative in both cases. This was not unexpected, given that the patient had no history of exposure to animal proteins in either a therapeutic or a social setting. The same sample was tested for RFs, using the local assay (Orgentec RF IgM), which returned a slightly elevated result. This slightly elevated concentration is not considered clinically significant, but may contribute to interference in the assay. Thus, heterophilic antibody interference remains the most likely cause of cTnI elevation in this patient. Further investigations have been limited, however, by recently decreasing concentrations of cTnI in this patient. Unpredictable variation in heterophilic antibody activity is a known phenomenon however, and the laboratory continues to oversee samples received from the patient.
Outcome and conclusions
The patient continues to regularly attend the Emergency Department with chest pain. No cardiac pathology has ever been identified in this patient and his chest pain has been attributed to a combination of gastro-oesophageal reflux disease and health anxiety. Senior review is now required in the Emergency Department before the patient is investigated or admitted. If ACS is ever suspected in this patient, the laboratory is able to pre-treat samples with heterophilic blocking tubes to provide an estimate of cTnI concentration, with referral for cTnT analysis required for a definitive result. This alternate pathway ensures that the patient is protected in case of a genuine cardiac event.
This case demonstrates that clinicians must be aware of assay interference, and highlights the benefit of discussing patients with the laboratory when test results do not correlate with clinical presentation. Laboratory staff members are rarely able to visit patients and, thus, are not in a position to suspect interference without input from the clinical team. In this case, failure to identify the underlying cause of elevated cTnI at an early stage resulted in a number of unnecessary and invasive investigations, significant costs to the NHS, and continued anxiety to the patient.
References
1. Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, et al. Third universal definition of myocardial infarction. Eur Heart J 2012; 33: 2551–2567.
2. National Institute of Health and Care Excellence (NICE). Chest pain of recent onset: assessment and diagnosis. NICE clinical guideline 95, 2010 (https://www.nice.org.uk/guidance/cg95).
3. Wu AHB. Analytical issues for clinical use of cardiac troponin. In: Morrow DA (ed.) Contemporary cardiology: cardiovascular biomarkers: pathophysiology and disease management. Humana 2006, pp 27–40.
4. Korff S, Katus HA, Giannitsis E. Differential diagnosis of elevated troponins. Heart 2006; 92: 987–993.
5. Plebani M. The detection and prevention of errors in laboratory medicine. Ann Clin Biochem 2010; 47: 101–110.
6. Sara JDS, Holmes DR, Jaffe AS. Fundamental concepts of effective troponin use: important principles for internists. Am J Med 2015; 128: 111–119.
7. Ver Elst KM, Spapen HD, Nguyen DN, Garbar C, Huyghens LP, Gorus FK. Cardiac troponins I and T are biological markers of left ventricular dysfunction in septic shock 2000; 46: 650–657.
8. Spies C, Haude V, Fitzner R, Schroder K, Overbeck M, Runkel N, Schaffartzik W. Serum cardiac troponin T as a prognostic marker in early sepsis. Chest 1998; 113: 1055–1063.
9. Jaffe AS. The 10 commandments of troponin with special reference to high sensitivity assays. Heart 2011; 97: 940–946.
10. Lippi G, Aloe R, Meschi T, Borghi L, Cervellin G. Interference from heterophilic antibodies in troponin testing. Case report and systematic review of the literature. Clin Chim Acta 2013; 426: 79–84.
11. Makaryus AN, Markaryus MN, Hassid B. Falsely elevated cardiac troponin I levels. Clin Cardiol 2007; 30: 92–94.
12. Zaidi A, Cowell R. False positive cardiac troponin elevation due to heterophile antibodies: more common than we recognise? BMJ Case Rep 2010; 2010: bcr1120092477.
13. Ismail AAA, Walker PL, Cawood ML, Barth JH. Interference in immunoassay is an underestimated problem. Ann Clin Biochem 2002; 39: 266–373.
14. Kaplan IV, Levinson SS. When is a heterophile antibody not a heterophile antibody? When it is an antibody against a specific immunogen. Clin Chem 1999; 45: 616–618.
The authors
Ceri Parfitt MSc, Christopher Duff* PhD
Department of Clinical Biochemistry, Pathology Directorate, Royal Stoke University Hospital, University Hospitals of North Midlands NHS Trust, Staffordshire, UK
*Corresponding author
E-mail: Chris.duff@uhnm.nhs.uk