Dilution testing as a novel alternative for confirmation of HIV rapid diagnostic testing in resource-limited settings
Rapid diagnostic testing enables life-saving scale up of HIV diagnosis but is vulnerable to false positive results. Confirmation testing can be impractical or cost prohibitive in resource-limited settings. Retesting a diluted blood sample is evaluated and proposed, at a proof of concept level, as a simple cost-effective HIV confirmation methodology.
by Derryck Klarkowski and Dr Erwan Piriou
Background
The diagnosis of HIV infection in developed countries is based on initial screening for HIV antibodies, and if detected, confirmation with nucleic acid testing (NAT) [1]. This ensures high sensitivity and specificity. However, the current World Health Organization (WHO) HIV Testing Services Guidelines do not include specific confirmation testing for the diagnosis of HIV across large population groups in resource-limited settings [2]. Instead WHO recommends that diagnosis be made on the basis of rapid diagnostic tests (RDTs) only (or equivalent enzyme immune assay tests) requiring a minimum of two positive test results, using test devices from different sources, for a positive diagnosis (or three in low prevalence settings) [2]. Although the WHO strategy has enabled life-saving scale-up of HIV diagnosis the significant compromise is that without confirmation there is a risk that patients/clients can be falsely diagnosed as HIV positive [3]. This is also well demonstrated in the study discussed now in this article where the WHO RDT algorithm resulted in 6.8% false positive results (n = 2897). Incorrect HIV diagnosis can have devastating consequences for the individual as well as wasting often over-stretched resources required for treatment and care.
Médecins Sans Frontières (MSF) has strongly advocated for the use of serological HIV confirmation testing in resource-limited settings when it is impractical to perform NAT [4–6]. Commercial confirmation kits are available that detect individual specific HIV antibodies, such as gp40, gp120, p24 and p32, that significantly increase the accuracy of testing at a considerably lower cost than NAT, and this type of confirmation testing can be performed by non-specialized laboratories. The downside, however, is that commercial confirmation kits nevertheless add cost, albeit reduced compared to NAT, and logistical complications that restrict their widespread use.
To address this, MSF has recently published a simplified confirmation approach based on antibody dilution requiring only the use of an additional routinely used RDT test device [7]. This study has been published as a ‘proof of concept’ paper and requires further testing across different settings for refinement before it can be generally recommended.
Causes of false positive HIV antibody detection
As with all tests, false positive HIV RDTs can be caused by user error (clerical mistakes, incorrect test performance, misinterpretation and cross-contamination). Other causes for HIV tests include nonspecific IgG binding [2], cross reactivity [2, 5], contaminating proteins [2] and pseudo-antigens created during the manufacturing process [5]. However, a key additional vulnerability for HIV antibody detection testing is that all commonly available HIV RDTs share a common gp41 detection antigen. Therefore, a cross-reactive antibody interacting with gp41 will act as a pan-cross-reactive antibody across multiple test devices [5].
The WHO algorithm is based on the assumption that HIV RDTs that use different antigen preparations are independent and, therefore, by requiring two positive tests (at a prevalence >5%) before reporting HIV positivity, the algorithm assumes that the second test confirms the result of the initial screening test [2]. However, in one MSF published study 50% of false positive samples had cross-reactive anti-gp41 activity, identified by Western blot (WB), that was the likely cause of the double false positive reactions with the two independent RDTs used in the testing algorithm [4].
MSF has proposed that early-immune-response broad-specificity polyclonal B-lymphocyte antibodies are a potential source of HIV RDT cross-reactive interfering antibodies [4]. These antibodies are likely to have increased frequency and intensity in resource-limited settings because of the higher prevalence of concomitant infections [8–10]. Additionally, displaced populations and individuals, such as caused by oppression, conflict and famine, are likely to have a greater vulnerability to cross-reacting infections than stable communities.
Theoretical basis for dilution methodology
Confirmation by dilution is based on the established sensitive/less sensitive (S/LS) methodology developed to identify recent HIV infection for the purposes of incidence surveys [11–15]. This methodology is based on the principle that HIV antibody titres increase over a period of several months after initial infection. Samples are initially tested using a high sensitivity HIV enzyme immunoassay (EIA) and if reactive are then further tested by the same EIA assay but using a diluted sample and reduced incubation time to reduce sensitivity. Samples testing positive on the sensitive (S) test but negative on the less sensitive (LS) test are designated a recent infection. The methodology has been successfully extended to the use of RDTs [13–15]. Confirmation by dilution adapts the S/LS principle to differentiate between high titre true HIV antibodies and low titre cross-reacting antibodies.
One postulated source of cross-reactive antibodies are broad spectrum antibodies produced in the early immune response to a wide range of infectious disease antigens, and these antibodies can cause nonspecific cross reactivity in HIV serological testing [5]. In proposing dilution as a methodology to confirm HIV infection, we postulate that cross-reacting antibodies will have a low titre relative to specific HIV antibodies.
Cross-reacting antibodies can generally be expected to have low avidity, as has been demonstrated by work in blood donors [16] and in MSF findings [4]. This will result in weakly positive results that can provide an alert for the tester; however, manufacturers generally state that any positive test line independent of strength should be interpreted as a positive result. Cross-reactive antibodies can also have high avidity as shown in a previous MSF publication where 7 of 24 (29.2%) false positive samples (total sample size 229) had strongly positive test lines in two RDTs but had a low titre relative to the confirmed true HIV antibodies [4].
The use of dilution as a supplementary confirmatory test by using antibody relative titres has been previously reported by Urwijitaroon et al. [17]. In another study, 41 samples were found positive using the HIV RDT Determine™ and 23 were negative on dilution [18]. Only 1 of these 23 samples was confirmed to be positive using serological confirmation (INNO-LIA™).
Field testing
A study was conducted at two sites in north western Ethiopia in programmes covering both residents and seasonal migrant workers. Seasonal workers are transient and, as postulated by MSF, may potentially have a higher risk of false positivity caused by cross-reacting antibodies [4, 5].
The study recruited 2897 individuals, and 265 (9.1%) samples tested as positive using two HIV RDTs from different manufacturers and would have been interpreted as HIV positive using the WHO algorithm [2]. Of the negative samples, 229 (approximately every 11th sample) were selected as a control. All algorithm-positive and negative control samples were further tested by dilution in situ, and additional confirmation testing performed by reference laboratories using WB and NAT for indeterminate WB samples.
All negative samples were confirmed as negative (100% sensitivity). However, 18/265 (6.8%) algorithm ‘positive’ samples were identified as HIV negative (false positive) by either WB or NAT.
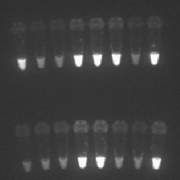
Dilution testing was performed by titrating the patient’s plasma using confirmed seronegative plasma from healthy blood donors using a micropipette. Ten microlitres of patient plasma was first diluted 1 : 10 in 90 µL of negative plasma followed by a serial 4-fold dilution from 1:40 to 1 : 10,240. Testing was performed using Determine™ HIV-1/2 (Alere Laboratories, Japan) following manufacturer’s instructions. Tests were interpreted as positive if there was any colouration of the test line and the highest dilution that gave a positive result was recorded. Where the lowest dilution (1 : 10) was negative, the sample was reported as negative.
Findings and conclusion
In this study, based on a specific population group over a specific time period, repeating the RDT test using the sample diluted 1 : 160 identified all false positive results and misidentified one true positive (see Table 1). However, there is a safety net that any sample with a reactive HIV RDT test that is not resolved as a true positive at the time of testing is not reported as negative but as inconclusive [2]. The patient/client is advised that testing has been inconclusive and testing should be repeated at a later time; WHO recommends retesting after 14 days. This allows time for true HIV antibodies to increase in titre.
The discriminatory threshold dilution may vary between different settings. In an earlier MSF study, a dilution of 1 : 1000 differentiated 229 true HIV positive from 27 HIV false positive samples (unpublished data, for further details see Klarkowski et al. [4]).
One strength of this MSF study is that NAT testing was available to resolve indeterminate WB samples which made it possible to rule out early seroconversion as a potential cause of false positive results. The limitation is that the findings are restricted to a single cohort with a single RDT and should be viewed as a ‘proof of concept’. More experience is needed in different settings and by different workers before the dilution methodology can be considered for potential scale up. It is proposed that the methodology has potential for use as a supplementary test in a confirmatory algorithm, whereby double positive RDT results are tested by dilution, with positive results above a determined threshold confirming HIV infection. Dilution results below the threshold would require further testing, such as repeat testing at a later time or NAT, to rule out false negative results either due to seroconversion or misclassification by the lower sensitivity dilution test.
References
1. Centers for Disease Control and Prevention and Association of Public Health Laboratories. Laboratory testing for the diagnosis of HIV infection: updated recommendations. 2014; http://stacks.cdc.gov/view/cdc/23447.
2. World Health Organization. Consolidated guidelines on HIV testing services. 2015; http://www.who.int/hiv/pub/guidelines/hiv-testing-services/en/.
3. Johnson C, Fonner V, Sands A, Tsui S, Ford N, Wong V, Obermeyer C, Baggaley R. Annex 14 A report on the misdiagnosis of HIV status. In: World Health Organization. Consolidated Guidelines on HIV Testing Services. 2015; http://www.who.int/hiv/pub/guidelines/hiv-testing-services/en/.
4. Klarkowski DB, Wazome JM, Lokuge KM, Shanks L, Mills CF, O’Brien DP. The evaluation of a rapid in situ HIV confirmation test in a programme with a high failure rate of the WHO HIV two-test diagnostic algorithm. PLoS One 2009; 4(2): e4351.
5. Klarkowski D, O’Brien DP, Shanks L, Singh KP. Causes of false positive HIV rapid diagnostic test results. Expert Rev Anti-infect Ther. 2013; 12(1): 49-62
6. Shanks L, Klarkowski D, O’Brien DP. False positive HIV diagnoses in resource limited settings: operational lessons learned for HIV programmes. PLoS ONE 2013; 8(3): e59906.
7. Shanks L, Siddiqui MR, Abebe A, Piriou E, Pearce N, Ariti C, Masiga J, Muluneh L, Wazome J, Ritmeijer K, Klarkowski D. Dilution testing using rapid diagnostic tests in a HIV diagnostic algorithm: a novel alternative for confirmation testing in resource limited settings. Virol J. 2015; 12: 75.DOI 10.1186/s12985-015-0306-4
8. Messele T, Abdulkadir M, Fontanet AL, Petros B, Hamann D, Koot M, Roos MT, Schellekens PT, Miedema F, Rinke de Wit TF. Reduced naive and increased activated CD4 and CD8 cells in healthy adult Ethiopians compared with their Dutch counterparts. Clin Exp Immunol. 1999; 115(3): 443–50.
9. Clerici M, Butto S, Lukwiya M, Saresella M, Declich S, Trabattoni D, Pastori C, Piconi S, Fracasso C, Fabiani M, Ferrante P, Rizzardini G, Lopalco L. Immune activation in Africa is environmentally-driven and is associated with upregulation of CCR5. Italian-Ugandan AIDS Project. AIDS 2000; 14(14): 2083–2092.
10. Clerici M, Declich S, Rizzardini G. African enigma: key player in human immunodeficiency virus pathogenesis in developing countries? Clin Diagn Lab Immunol. 2001; 8(5): 864–866.
11. World Health Organization Technical Working Group on HIV Incidence Assays. When and how to use assays for recent infection to estimate HIV incidence at a population level. 2011; http://www.who.int/diagnostics_laboratory/hiv_incidence_may13_final.pdf 2011.
12. Constantine NT, Sill AM, Jack N, Kreisel K, Edwards J, Cafarella T, Smith H, Bartholomew C, Cleghorn FR, Blattner WA. Improved classification of recent HIV-1 infection by employing a two-stage sensitive/less-sensitive test strategy. J Acquir Immune Defic Syndr. 2003; 32: 94–103.
13. Soroka SD, Granade TC, Candal D, Parekh BS. Modification of rapid human immunodeficiency virus (HIV) antibody assay protocols for detecting recent HIV seroconversion. Clin Diagn Lab Immunol. 2005; 12: 918–21.
14. Kshatriya R, Cachafeiro AA, Kerr RJS, Nelson JA, Fiscus SA. Comparison of two rapid human immunodeficiency virus (HIV) assays, Determine™ HIV-1/2 and OraQuick Advance Rapid HIV-1/2, for detection of recent HIV seroconversion. J Clin Microbiol. 2008; 46(10): 3482–3483.
15. Girardi SB, Barreto AM, Barreto CC, Proietti AB, Carvalho SM, Loureiro P, Sabino EC. Evaluation of rapid tests for human immunodeficiency virus as a tool to detect recent seroconversion. Braz J Infect Dis. 2012; 16(5): 452–456.
16. Bouillon M, Aubin E, Roberge C, Bazin R, Lemieux R. Reduced frequency of blood donors with false-positive HIV-1 and -2 antibody EIA reactivity after elution of low-affinity nonspecific natural antibodies. Transfusion 2002; 42(8): 1046–1052.
17. Urwijitaroon Y, Barusrux S, Romphruk A, Puapairoj C, Thongkrajai P. Anti-HIV Antibody Titer: An Alternative Supplementary Test for Diagnosis of HIV-1 Infection. Asian Pac J Allergy Immunol. 1997; 15:193–198.
18. Duedu KO, Hayford AA and Sagoe KW. Misclassification of recent HIV-1 seroconversion in sub-Saharan Africa using the sensitive/less sensitive technique. Virol J. 2011; 8: 176.
The authors
Derryck Klarkowski* MAppSc, Erwan Piriou PhD
Médecins Sans Frontières, Amsterdam, The Netherlands
*Corresponding author
E-mail: derryck.klarkowski@gmail.com