G protein-coupled receptors, accessory proteins and signalling
Molecular diagnostics is increasingly embracing pharmacogenomics. Here we discuss the role of G protein-coupled receptors and their accessory proteins in disease, drawing on our experience addressing the role of the calcium-sensing receptor polymorphisms/variation in familial hypocalciuric hypercalcemia and autosomal dominant hypocalcemia in order to highlight the role that pharmacogenomics may play in personalized treatment.
by Dr M. D. Thompson, Dr D. E. C. Cole and Dr G. N. Hendy
Introduction
The identification and characterization of gene families encoding G protein-coupled receptors (GPCRs) and the proteins necessary for the processes of ligand binding, GPCR activation, inactivation and receptor trafficking facilitates the study of drug response in the context of human genetic disease. Thompson et al. reviewed these topics in Volume 1175 of Methods in Molecular Biology in 2014 [1–3].
With the advance of genomic technologies, there has been a substantial increase in the inventory of naturally occurring rare and common GPCR variants [2, 3]. In addition to functional GPCR variants, genetic variation has been found in a variety of G protein subunits and accessory proteins that normally modify or organize heterotrimeric G protein coupling. These include variants of the regulator of G protein signalling (RGS) protein associated with hypertension; variants of the activator of G protein signalling (AGS) proteins associated with various phenotypes (such as the type III AGS8 variant to hypoxia); variants in of the G protein-coupled receptor kinase (GRK) proteins, such as GRK4, associated with disorders such as hypertension [1]. Variation in GPCR, G protein and accessory protein structure and function provides the basis for examining the pharmacogenomics of GPCRs and the genetics of related monogenic disorders [1–3].
GPCR variants and variant G protein subunits associated with human disease
Diseases caused by the genetic disruption of GPCR functions may be selectively targeted by drugs that rescue altered receptors. The identification of variants in these receptors provides genetic reagents useful in drug screens. Examples of drugs developed as a result of targeting GPCRs mutated in disease include: the calcimimetics and calcilytics, drugs targeting melanocortin receptors in obesity and interventions that alter gonadotropin-releasing hormone receptor (GNRHR) loss from the cell surface in idiopathic hypogonadotropic hypogonadism [2, 3].
Inactive, overactive and constitutively active receptors
Genetic variations in GPCR genes disrupt GPCR function in a variety of human genetic diseases. In vitro studies and animal models have been used to identify the molecular pathologies underlying these GPCR mutations. Inactive, overactive, or constitutively active receptors have been identified. These receptor variants alter ligand binding, G protein coupling, receptor desensitization, or receptor recycling. Variant GPCRs disrupted in disease include rhodopsin, thyrotropin, parathyroid hormone (PTH), melanocortin, follicle-stimulating hormone (FSH), luteinizing hormone, GNRHR, adrenocorticotropic hormone, vasopressin, endothelin-β, purinergic, and the G protein associated with asthma [GPRA or neuropeptide S receptor 1 (NPSR1)] [2]. Data on the role of activating and inactivating calcium-sensing receptor (CASR) mutations provide examples that will be discussed in detail with respect to familial hypocalciuric hypercalcemia (FHH) and autosomal dominant hypocalcemia (ADH) [4].
Calcium-sensing receptor mutations and hypercalcemia/hypocalcemia
The CASR functions as an extracellular calcium sensor for the parathyroid gland and the kidney. The CASR itself is a plasma membrane GPCR that is abundantly expressed in the PTH secreting cells of the parathyroid gland and the cells lining the renal tubule lumen [2, 4]. The activity and/or expression levels of the CASR dictate the calcium set-point at which PTH is secreted from the parathyroid gland [2]. CASR gene variants may influence many physiological processes by contributing to individual differences in calcium metabolism [2].
Inherited abnormalities of the CASR gene give rise to a variety of disorders of mineral ion homeostasis [5]. Heterozygous loss-of-function mutations cause familial (benign) hypocalciuric hypercalcemia (FHH) in which the lifelong mild hypercalcemia is generally asymptomatic. Homozygous inactivating mutations give rise to neonatal severe hyperparathyroidism (NSHPT) with extreme hypercalcemia and marked skeletal changes [5–7]. Heterozygous activating mutations of the CASR cause ADH that may be asymptomatic or present with seizures in the neonatal period or childhood or later in life [2].
Familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism
The syndrome known as familial hypocalciuric hypercalcemia (FHH), or familial benign hypercalcemia, results in mild primary hyperparathyroidism and relatively normal serum concentrations of PTH [8]. A key feature of FHH is the unusually high renal tubular reabsorption of calcium and magnesium in the face of hypercalcemia. However, some FHH families have affected members in which calcium excretion is increased and this may reflect the particular CASR mutation involved [2].
NSHPT involves multiglandular parathyroid hyperplasia. Children under the age of 6 months develop severe, symptomatic hypercalcemia with bony changes of hyperparathyroidism. Delay in treatment can lead to a devastating neurodevelopmental disorder. Some forms of neonatal hyperparathyroidism, involving either a de novo or paternal inheritance of a mutated CASR allele, present with milder symptoms [2].
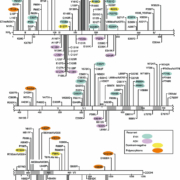
Upwards of 200 unique inactivating, FHH/NSHPT type mutations in the CASR have been identified [2], as shown in Figure 1 (http://www.casrdb.mcgill.ca/). Although FHH is inherited in an autosomal dominant manner with almost 100% penetrance and variable expressivity, the population prevalence is not well defined. The FHH trait was initially mapped to chromosome 3q21, the locus of the CASR gene: two-thirds of FHH cases are due to mutations in the CASR gene and the disorder is FHH type 1 [2].
In some kindreds, however, the FHH trait maps to either chromosome 19p13.3 (FHH type 2) or 19q13.3 (FHH type 3). FHH2 is due to heterozygous loss-of-function mutations in GNA11, the gene encoding the alpha subunit of G11 that couples the activated CASR to intracellular signalling pathways [9]. FHH3 is due to inactivating mutations in the AP2S1 gene that encodes the sigma subunit of adaptor protein complex 2 critical for clathrin-mediated endocytosis of a variety of cell surface proteins including GPCRs such as the CASR [2].
Hypocalcemia, hypoparathyroidism, and hypocalcemic hypercalciuria
Gain-of-function mutations in the CASR gene have been identified in several families previously diagnosed with ADH, autosomal dominant hypoparathyroidism, and hypocalcemic hypercalciuria [2]. In the parathyroid gland, the activated CASR suppresses PTH secretion and in the kidney, it induces hypercalciuria [4]. De novo mutations are common [2]. Mosaicism for de novo mutation in an otherwise healthy parent has been described and this has important implications for counselling parents about the risk of recurrence [2].
In a subset of ADH families, CASR gain-of-function mutations have been associated with the onset of tonic–clonic seizures. In ADH, brain calcifications – sometimes accompanied by seizures – suggest that activating mutations may alter calcium homeostasis in the brain. The abnormal set-point of calcium regulation complicates treatment with calcitriol and dietary calcium supplementation because the CASR expressed in the kidney may override other regulators of calcium excretion. The constitutively activated CASR mutant induces hypercalciuria, which may exacerbate the hypocalcemia [2, 10].
More than 100 activating mutations (virtually all missense) have been identified and appear almost equally divided between the amino-terminal third of the extracellular domain (ECD) and the transmembrane domain shown in Figure 1 (http://www.casrdb.mcgill.ca/).
GPCR pharmacogenomics
Pharmacogenetics investigates the influence of genetic variants on physiological phenotypes related to drug response and disease, while pharmacogenomics takes a genome-wide approach to advancing this knowledge. Both play an important role in identifying responders and non-responders to medication, avoiding adverse drug reactions, and optimizing drug dose for the individual.
The CASR provides an example of GPCR variability in the population. While CASR variants contribute to monogenic disorders such as FHH and ADH, common CASR polymorphisms also account for some of the population variation in calcium response that is a risk factor for a variety of disease susceptibilities. CASR single nucleotide polymorphisms (SNPs) have been associated with a number of complex phenotypes. For example, the Ala986Ser variant may contribute to bone mineral density, primary hyperparathyroidism, and Paget disease [11].
The cluster of missense polymorphisms located in the cytoplasmic tail of the receptor is associated with inter-individual population differences in Ca2+ metabolism [12]. Different haplotypes are associated with primary hyperparathyroidism and the frequency of kidney stones. More recent genome-wide association studies in ~33,000 individuals of European and Indian Asian ancestry confirmed that the blood calcium concentration associated most significantly with SNPs in the CASR gene [13].
CASR variants are known to alter the sensitivity of the CASR and result in altered extracellular calcium-concentration set points in tissues. Web sites such as http://www.casrdb.mcgill.ca/ document a number of SNPs scattered across the more than 100 kb region of genomic DNA that encompasses the CASR gene. Common missense SNPs (Ala986Ser and Arg990Gly) are clustered in the DNA region encoding the cytoplasmic tail of CASR. The most common of these, the Ala986Ser variant, is predictive of the unbound, extracellular calcium levels [11]. The Ala986Ser variant is thus a mild inactivating variant that may predispose to hypercalcemia without being fully predictive of hypocalciuria. By contrast, the Arg990Gly variant (activating) results in the increased calcium excretion that characterizes idiopathic hypercalciuria and is predictive of nephrolithiasis [2].
Conclusion
GPCRs are the primary target of therapeutic drugs and have been the focus of these studies. These variants include SNPs and insertion/deletions that have potential to alter GPCR expression of function. In vivo and in vitro studies have determined functional roles for many GPCR variants, but genetic association studies that define the physiological impact of the majority of these common variants are still limited. Despite the breadth of pharmacogenetic data available, GPCR variants have not been included in drug labelling and are only occasionally considered in optimizing clinical use of GPCR targeted agents. As the extent of GPCR pharmacogenomic data increases, the opportunity for routine assessment of GPCR variants to predict disease risk, drug response and potential adverse drug effects will no doubt become more commonplace.
References
1. Thompson MD, Cole DE, Jose PA, et al. G protein-coupled receptor accessory proteins and signaling: pharmacogenomic insights. Methods Mol Biol. 2014; 1175: 121-52.
2. Thompson MD, Hendy GN, Percy ME, et al. G protein-coupled receptor mutations and human genetic disease. Methods Mol Biol. 2014; 1175: 153-87.
3. Thompson MD, Cole DE, Capra V, et al. Pharmacogenetics of the G protein-coupled receptors. Methods Mol Biol. 2014; 1175: 189-242.
4. Zhang C, Zhuo Y, Moniz HA, et al. Direct determination of multiple ligand interactions with the extracellular domain of the calcium sensing receptor. J Biol Chem. 2014 Oct 10. pii: jbc.M114.604652.
5. Thim SB, Birkebaek NH, et al. Activating calcium-sensing receptor gene variants in children: a case study of infant hypocalcaemia and literature review. Acta Paediatr. 2014 Jul 10. doi: 10.1111/apa.12743.
6. Toka HR, Pollak MR.The role of the calcium-sensing receptor in disorders of abnormal calcium handling and cardiovascular disease. Curr Opin Nephrol Hypertens. 2014; 23: 494-501.
7. Grzegorzewska AE, Ostromecki G. Gene polymorphism of the vitamin D receptor, vitamin D-binding protein and calcium-sensing receptor in respect of calcium-phosphate disturbances in chronic dialysis patients. Przegl Lek. 2013; 70: 735-8.
8. Jakobsen NF, Rolighed L, Nissen PH, et al. Muscle function and quality of life are not impaired in familial hypocalciuric hypercalcemia: a cross- sectional study on physiological effects of inactivating variants in the calcium-sensing receptor gene (CASR). Eur J Endocrinol. 2013; 169: 349-57.
9. Nesbit MA, Hannan FM, Howles SA, et al. Mutations affecting G-protein subunit α11 in hypercalcemia and hypocalcemia. N Engl J Med. 2013; 368: 2476-86.
10. Ranieri M, Tamma G, Di Mise A, et al. Excessive signal transduction of gain-of-function variants of the calcium-sensing receptor (CaSR) are associated with increased ER to cytosol calcium gradient. PLoS One. 2013; 8: e79113.
11. Han G, Wang O, Nie M, et al. Clinical phenotypes of Chinese primary hyperparathyroidism patients are associated with the calcium-sensing receptor gene R990G polymorphism. Eur J Endocrinol. 2013; 169: 629-38.
12. Scillitani A, Guarnieri V, Battista C, et al. Primary hyperparathyroidism and the presence of kidney stones are associated with different haplotypes of the calcium-sensing receptor. J Clin Endocrinol Metab. 2007; 92: 277-83.
13. Kapur K1, Johnson T, Beckmann ND, et al. Genome-wide meta-analysis for serum calcium identifies significantly associated SNPs near the calcium-sensing receptor (CASR) gene. PLoS Genet. 2010; 6: e1001035.
The authors
Miles D. Thompson1* PhD; David E. C. Cole2 MD, PhD; Geoffrey N. Hendy3 PhD
1Department of Pharmacology and Toxicology, Medical Sciences Building, University of Toronto, Toronto, ON, Canada. M5S 1A8.
2Departments of Laboratory Medicine and Pathobiology, Medicine and Genetics, University of Toronto, ON, Canada. M4N 3M5.
3Departments of Medicine, Physiology, and Human Genetics, McGill University, and Calcium Research Laboratory and Hormones and Cancer Unit, Royal Victoria Hospital, Montreal, QC, Canada. H3A 1A1.
*Corresponding author
E-mail: miles.thompson@utoronto.ca